Data summary & Data processing
Data summary
Beyond of adding new created small variations from the databases of NCBI dbSNP, new types of regulatory elements and more extend annotation (such as SNP related disease), significant adjustments on database structure and function have been made in this update. The concerns of rSNPBase 3.0 were extended from regulatory elements that position overlap SNPs to SNP related regulatory element-target gene (E-G) pairs. Based on the annotation of regulatory element-gene pairs, rSNPBase 3.0 supports SNP-based regulatory network analysis. A summarized data content of rSNPBase 3.0 is shown in Table 1.
Data processing
As shown in Figure 1, several types of regulatory elements were gotten from reference databases (mostly of them provide experimental supported data), their relation with genes from Ensembl (GRCh37) were analyzed with genomic proximity or by using reference databases. Genome-wide human SNPs from NCBI dbSNP (build 150) were analyzed and filtered by involved regulatory elements sets and thus connected to corresponding E-G pairs in the same time. The analysis results were stored in rSNPBase 3.0 and presented as regulatory SNP (rSNP) reports and SNP-based regulatory network.
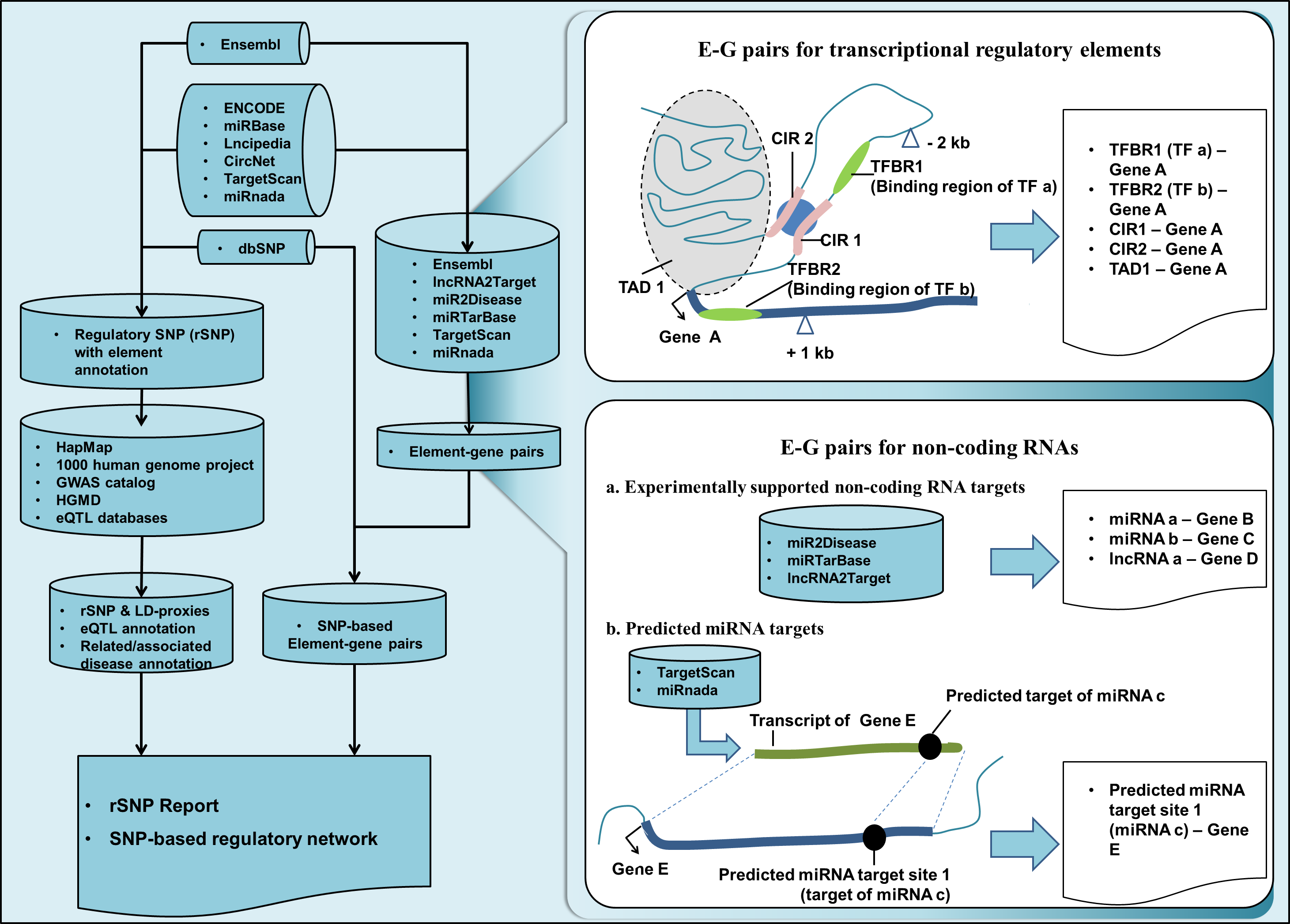
Figure 1. Data processing and data content of rSNPBase 3.0
Reference data
References
1. Mathelier, A., Zhao, X., Zhang, A.W., Parcy, F., Worsley-Hunt, R., Arenillas, D.J., Buchman, S., Chen, C.Y., Chou, A., Ienasescu, H. et al. (2014) JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic acids research, 42, D142-147.
2. Kheradpour, P. and Kellis, M. (2014) Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic acids research, 42, 2976-2987.
3. Rosenbloom, K.R., Armstrong, J., Barber, G.P., Casper, J., Clawson, H., Diekhans, M., Dreszer, T.R., Fujita, P.A., Guruvadoo, L., Haeussler, M. et al. (2015) The UCSC Genome Browser database: 2015 update. Nucleic acids research, 43, D670-681.
4. Fu, Y. and Weng, Z. (2005) Improvement of TRANSFAC matrices using multiple local alignment of transcription factor binding site sequences. Genome informatics. International Conference on Genome Informatics, 16, 68-72.
5. Kozomara, A. and Griffiths-Jones, S. (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic acids research, 42, D68-D73.
6. Friedman, R.C., Farh, K.K., Burge, C.B. and Bartel, D.P. (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome research, 19, 92-105.
7. Betel, D., Wilson, M., Gabow, A., Marks, D.S. and Sander, C. (2008) The microRNA.org resource: targets and expression. Nucleic acids research, 36, D149-153.
8. Volders, P.J., Helsens, K., Wang, X.W., Menten, B., Martens, L., Gevaert, K., Vandesompele, J. and Mestdagh, P. (2013) LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic acids research, 41, D246-D251.
9. Liu, Y.C., Li, J.R., Sun, C.H., Andrews, E., Chao, R.F., Lin, F.M., Weng, S.L., Hsu, S.D., Huang, C.C., Cheng, C. et al. (2016) CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res, 44, D209-215.
10. Jiang, Q., Wang, Y., Hao, Y., Juan, L., Teng, M., Zhang, X., Li, M., Wang, G. and Liu, Y. (2009) miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic acids research, 37, D98-104.
11. Hsu, S.D., Lin, F.M., Wu, W.Y., Liang, C., Huang, W.C., Chan, W.L., Tsai, W.T., Chen, G.Z., Lee, C.J., Chiu, C.M. et al. (2011) miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic acids research, 39, D163-169.
12. Jiang, Q., Wang, J., Wu, X., Ma, R., Zhang, T., Jin, S., Han, Z., Tan, R., Peng, J., Liu, G. et al. (2015) LncRNA2Target: a database for differentially expressed genes after lncRNA knockdown or overexpression. Nucleic Acids Res, 43, D193-196.
13. Welter, D., MacArthur, J., Morales, J., Burdett, T., Hall, P., Junkins, H., Klemm, A., Flicek, P., Manolio, T., Hindorff, L. et al. (2014) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic acids research, 42, D1001-1006.
14. Stenson, P.D., Ball, E.V., Mort, M., Phillips, A.D., Shaw, K. and Cooper, D.N. (2012) The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr Protoc Bioinformatics, Chapter 1, Unit1 13.
15. Xia, K., Shabalin, A.A., Huang, S., Madar, V., Zhou, Y.H., Wang, W., Zou, F., Sun, W., Sullivan, P.F. and Wright, F.A. (2012) seeQTL: a searchable database for human eQTLs. Bioinformatics, 28, 451-452.
16. Gamazon, E.R., Zhang, W., Konkashbaev, A., Duan, S., Kistner, E.O., Nicolae, D.L., Dolan, M.E. and Cox, N.J. (2010) SCAN: SNP and copy number annotation. Bioinformatics, 26, 259-262.
17. Cline, M.S., Craft, B., Swatloski, T., Goldman, M., Ma, S., Haussler, D. and Zhu, J. (2013) Exploring TCGA Pan-Cancer data at the UCSC Cancer Genomics Browser. Scientific reports, 3, 2652.
18. Schadt, E.E., Molony, C., Chudin, E., Hao, K., Yang, X., Lum, P.Y., Kasarskis, A., Zhang, B., Wang, S., Suver, C. et al. (2008) Mapping the genetic architecture of gene expression in human liver. PLoS biology, 6, e107.
19. Myers, A.J., Gibbs, J.R., Webster, J.A., Rohrer, K., Zhao, A., Marlowe, L., Kaleem, M., Leung, D., Bryden, L., Nath, P. et al. (2007) A survey of genetic human cortical gene expression. Nature genetics, 39, 1494-1499.
20. Stranger, B.E., Nica, A.C., Forrest, M.S., Dimas, A., Bird, C.P., Beazley, C., Ingle, C.E., Dunning, M., Flicek, P., Koller, D. et al. (2007) Population genomics of human gene expression. Nature genetics, 39, 1217-1224.
21. Barski, A., Cuddapah, S., Cui, K., Roh, T.Y., Schones, D.E., Wang, Z., Wei, G., Chepelev, I. and Zhao, K. (2007) High-resolution profiling of histone methylations in the human genome. Cell, 129, 823-837.
22. Pickrell, J.K., Marioni, J.C., Pai, A.A., Degner, J.F., Engelhardt, B.E., Nkadori, E., Veyrieras, J.B., Stephens, M., Gilad, Y. and Pritchard, J.K. (2010) Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature, 464, 768-772.
23. Montgomery, S.B., Sammeth, M., Gutierrez-Arcelus, M., Lach, R.P., Ingle, C., Nisbett, J., Guigo, R. and Dermitzakis, E.T. (2010) Transcriptome genetics using second generation sequencing in a Caucasian population. Nature, 464, 773-777.
24. Zeller, T., Wild, P., Szymczak, S., Rotival, M., Schillert, A., Castagne, R., Maouche, S., Germain, M., Lackner, K., Rossmann, H. et al. (2010) Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PloS one, 5, e10693.
25. Ma, B., Huang, J. and Liang, L. (2014) RTeQTL: Real-Time Online Engine for Expression Quantitative Trait Loci Analyses. Database : the journal of biological databases and curation, 2014.
26. Ramasamy, A., Trabzuni, D., Guelfi, S., Varghese, V., Smith, C., Walker, R., De, T., Consortium, U.K.B.E., North American Brain Expression, C., Coin, L. et al. (2014) Genetic variability in the regulation of gene expression in ten regions of the human brain. Nature neuroscience, 17, 1418-1428.
27. Ding, J., Gudjonsson, J.E., Liang, L., Stuart, P.E., Li, Y., Chen, W., Weichenthal, M., Ellinghaus, E., Franke, A., Cookson, W. et al. (2010) Gene expression in skin and lymphoblastoid cells: Refined statistical method reveals extensive overlap in cis-eQTL signals. American journal of human genetics, 87, 779-789.
28. Consortium, G.T. (2015) Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science, 348, 648-660.
29. Abecasis, G.R., Altshuler, D., Auton, A., Brooks, L.D., Durbin, R.M., Gibbs, R.A., Hurles, M.E. and McVean, G.A. (2010) A map of human genome variation from population-scale sequencing. Nature, 467, 1061-1073.
30. Patterson, K. (2011) 1000 genomes: a world of variation. Circulation research, 108, 534-536.
31. Li, Y., Willer, C.J., Ding, J., Scheet, P. and Abecasis, G.R. (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genetic epidemiology, 34, 816-834.

